

Discussed below is the case of cooperative binding of a substrate to a multisubunit enzyme. This is placed here as an introduction to "allosteric bonding". Instead of a compound binding to a second site, it involves only substrate binding to an active site. BUT the priciples of structural rearrangement discussed here also apply to all the regulation mechanisms mentioned on the previous page. It also introduces a protein (hemogolbin) that is the paradigm for these ideas.
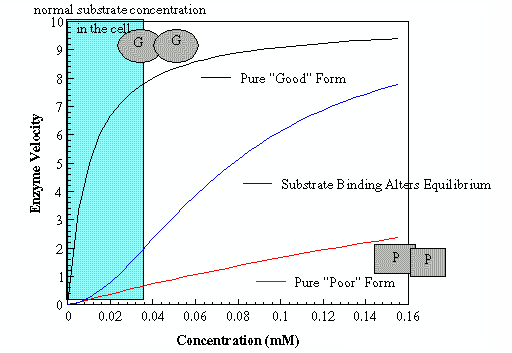
Some enzymes do not behave according the simple Michaelis - Menten kinetics scheme that was derived in module 5, but rather have a very complex sigmoidal response when you plot substrate concentration [S] vs. vobs. Like the Blue line in the graph to the right.
In order to explain this more complex curve, We must look ar protein structure one more time. Enzymes that exhibit this behaviour contain more than one subunit and generally the subunits are identical (or least very similar) in sequence and structure. So far, protein structure has been discussed as if it is a static structure or at least required covalent modification such as an amide bond cleavage to alter structure appreciably. That model has been sufficient up to now to demonstrate the principles of catalys. Now we need to re-examine this a little bit.
How is it the efficiency of the enzyme changes in "midstream" as [S] increases? This is most easily explained by invoking two populations of structures one that works well (high efficiency or GOOD catalytic properties) and one that does not work so well (low efficiency or POOR catalytic properties) Note: The biochemistry world since the beginning of the theory of cooperativity has called the what I call the GOOD form - R (for relaxed) and the POOR form as T (for tense). I am forever getting the R/T nomenclauture confused. I am not the only one I have seen text books with blatant errors using the R/T convention only adding to the confusion. I will use GOOD (R) and POOR (T) nomenclature throughout this module
Both structural forms are pesent at all times. There is an equilibrium between them that does not depend on catalysis or another protein or any other outside interference. Rather the two forms switch between them in a rapid and facile manner. The forces involved in this process are the same as those invovled in maintenance of all protein structures but complicated by remembering that atoms of a molecule/protein are not static but are in constant motion and vibration.
Furthermore, multisubunit proteins like to maintain symmetry (just the nature of interactions between subunits). Therefore, if the structure of one subunit in a protein is in the POOR (T) form then all of the subunits within that protein molecule are in the POOR (T) form. Like wise for the GOOD (R) form. If one subunit within a protein molecule is GOOD then all are GOOD within that molecule. There is, however, no influence on structure between molecules. As in the dimeric protein represented by the circles and squares in the picture, there are no proteins that have one POOR and one GOOD.
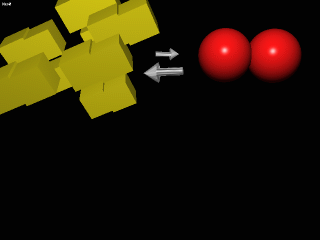 |
In order for these complicated kinetics to be observed it is important to know that the two structures (POOR and GOOD) are in equilibrium. In the absence of [S] This equilibrium lies far toward the POOR structural form of the protein. This does NOT mean that there in NO GOOD structures.... just not very many. As S is added to the solution it will preferentially bind to the few GOOD structures that are available, because this is the form that has the much lower Km. Put another way the GOOD form is better optimized for binding and catalysis of S. But with the very act of binding S the form has changed to GOOD·S. Thus lowering the concentration of the GOOD form (no S bound). By the principles of equilibrium some of the POOR form must switch to the GOOD form (no S bound). The net effect of S binding is to lower the concetration of POOR enzyme form in solution.
It is important to note here that it is NOT S binding to the protein that causes a structural change but rather S binding causes a shift in the equilibrium!