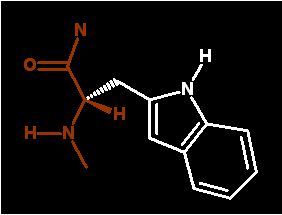
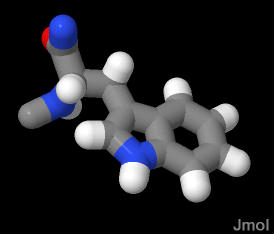

very large bulky ring structure sidechain that is pretty hydrophobic and aromatic
Almost all proteins absorb light in the ultraviolet region (not vivble to humans) at 280 nm due the presence of aromatic amino acids tyrosine and tryptophan.
Due to the large aromatic ring structure this side chain has a large light absorption property at 280nm. On a molar basis about 4 times higher than that of tyr. But since there is (on average) less TRP than TYR accounts for less of the total absorption for proteins.
Aromatic side chain. The Nitrogen in the ring gives it some polarity, In deed, the nitorgen can act as a hydrogen bond donor under some circumstances.
In the demonstration below, the TRP is in a section of random coil. Most of it is buried in the protein structure sequestered away from water, however the nitrogen if the ring is peeking out into the water.
  Atom Label Description | |
|
Click an atom to diplay it's identity here | |
|
Messages about the currently highlighted features |