This is the dominant form
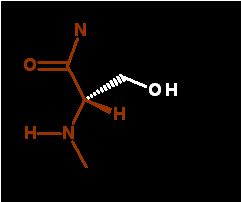
There is very little of this under normal conditions
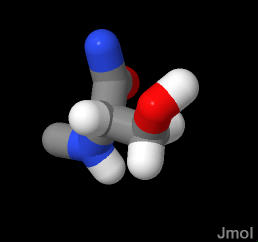

Polar amino acid can act as a hydrogen bond donor or acceptor. This is its most frequent role in structure. The pK of about 15 for the sidechain -OH is way above the physiological range. Normally deprotonation of this group can be ignored.
However, there are some exceptions that we will run into in this course. In the serine proteases (blood clotting enzymes) a serine is very close to a HIS in the active site. This special case has the effect of greatly lowering the pK and allowing it to deprotonate under physiological conditions. More on this in later modules
In the demonstration below, the SER is in a β-sheet and is exposed to water. It is safe to assume in this NORMAL case that the serine has a normal pK and is protonated and neutral.
  Atom Label Description | |
|
Click an atom to diplay it's identity here | |
|
Messages about the currently highlighted features |