|
|
|
| Name | 3 Letter | 1 Letter |
Lysine |
LYS |
K |
|
Unprotonated (base form - neutral)
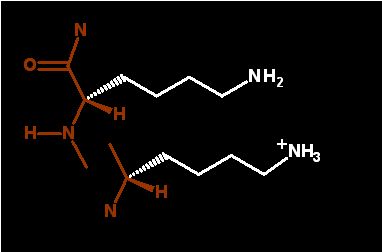
Protonated (acid form - positively charged)
dominant in water |
 |

 |
|
Drawn as if part of protein to emphasize the sidechain properties
| |
The "Sidechain Polarity" button draws an an envelope around the sidechain that is colored by charge according to the scale above. |
|
| pKR in H2O= 9 |
Free amino acid Mass = 146 g/M |
|
| Probability of being found in a: |
| α-Helix |
β-Sheet |
β-Turn |
| 75% |
50% |
60% |
| Special Attributes |
|
This amino acid sidechain ends with a primary amine which is generally protonated and ionized (positively) at physiological pH.
The charge means that it is usually on the surface of a water soluble protein where it is exposed to water
In the demonstration below the lysine is at the beginning of an α-helix and interestingly the terminal amine group in this particular case does not appear to be sticking out into water - but rather looks to be buried in the protein... such is the way
|
In the demonstration below you may "left click and drag" to rotate the molecule "SHIFT left click and drag (up or down)" to make smaller or bigger
Click on the white squares in succession to turn or/off identifying features. Some text describing the issues is in the lower right text box |
|
|


Atom Label Description

|
|
Click an atom to diplay it's identity here
|
|
Messages about the currently highlighted features
|
|
| Jmol: an open-source Java viewer for chemical structures in 3D. http://www.jmol.org/ |
|





