
Elements of Protein Structure
Inter Sidechain Covalent Bonds
So far we have discussed only non-covalent interactions between sidechain atoms in a protein structure. The sidechains of Cys are capable of generating a covalent bond between them ONCE the protein has folded. IF two cysteines happen to be near each other after the final structure has formed then the two sulfur atoms that are near each other can each get OXIDIZED to form sulfur-sulfur covalent bond, called a disulfide bond. The covalent bond is many times stronger than ANY of the non-covalent interactions discussed thus far. Once formed, a disulfide bond has a great deal of influence in maintaining the structure of the protein.
Notice that this is a oxidation reaction.... There is a NET LOSS of two electrons from the 2 sulfur atoms (and two protons as well) as the bond forms. In order to break this bond again the disulfide bond must be reduced. In the test tube. there are a couple reagents commonly available for this reduction reaction: dithiothreitol (DTT) and β-mercaptoethanol (βME). (The latter smells STRONGLY of rotten eggs).
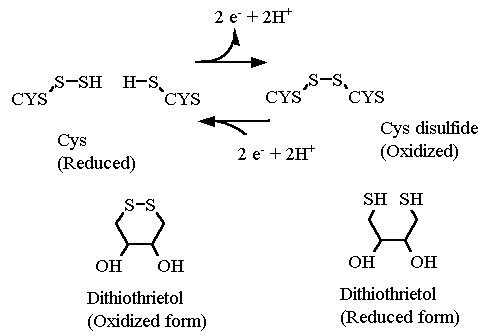
The formation of these disulfide bonds occurs mostly in proteins that are excreted from a cell. For example, many proteins of the blood coagulation pathway, which are found in the blood serum and not inside a cell, have disulfide bonds in their structure. Most proteins inside a cell however, while they likely contain Cysteines contain almost no disulfide bonds. This is because the cell cytosol is a very reducing environment and there are many enzyme systems (thioredoxins) devoted to maintaining cysteiens in the reduced state.
I have made a big deal earlier about the protein structure forming before the disulfide bond forms. The formation of the disulfide does not in any way determine the final protein structure. It simply makes it MUCH more difficult to unfold the protein after it has formed.
Disulfide bonds and superglue
Much like putting to a broken china plate back together with super glue. Even without the glue, the pieces of the plate fit together well, and may even stick together some. But upon applying glue and letting it harden it is much more difficult to get the pieces back apart again. Note: the glue does not determine HOW the pieces fit together, it merely holds them together once the pieces are properly assembled. If you applied the glue without knowing how the pieces fit first, in your hurry to put it together before the glue hardened you might end up attaching incorrect pieces to each other ... ending up with a nonfunctional plate.
The importance of the correct sequence for folding and disulfide bond formation
The correct order.. and how it happens in living tissue as well
- Proper protein folding
- Disulfide formation
This can be demonstrated with a classic experiment with RNase. This experiment is the topic of two of the discussion questions for this module. The protein folding and disulfide bond formation can be experimentally controlled independently in a test tube. When the protein is purified (more about this in the next module) it is an active catalyst and is known to have four disulfide bonds amongst the eight cysteines in the sequence.
A compound called urea has a tendency to make proteins unravel from their normal folded and active state to one that is called denatured. Denatured proteins do not have a single structure but rather many, many that are random AND rapidly fluctuating. For RNAse, urea alone cannot do the trick because of the four disulfide bonds. Therefore DTT must also be added to reduce the disulfides so that the protein can completely denature.
After being completely denatured, RNAse can be "refolded" in two ways:
- refolding and then formation of disulfide bonds (remove urea and then the DTT)
- formation of disulfide bonds and then attempting to refold (remove DTT and then the urea)
Perfomed in the correct sequence in one test tube experiment, mostly active enzyme results. Performed in the "incorrect" sequence in a test tube experiment mostly inactive enzyme results. I have given you half the answer... I'll let you figure out which is the correct order (should be obviuos from the first part of this page) and why the incorrect sequence results in inactive (unproperly folded) RNAse.
| |
| Below is a model of Factor VIIa (the active form of Factor VII the blood clotting cascade). This protein does contain two subunits. However, they are made a little differently than indicated in the previous page. Here a single polypeptide (factor VII) is synthesized in the cell and then excreted into the blood stream. It is "activated" by the cleavage of single amide bond clipping it into two separate pieces called "heavy" and "light". They stay associated. You can also show all the Cysteine amino acids in the protein. There are 16 in total and they form 8 disulfide bonds. See if you can explain why the heavy and light chains always stay associated.
If you hover the mouse over a cysteine you can see where in the sequence it is. Notice how some cysteines can be very far apart in sequence but close in space.
Reiteration... Cysteines must be close in space before they can form a disulfide bond.
|
|
|
|

|
|
Click an atom to diplay it's identity here
|
|
Messages about the currently highlighted features
|
rotate molecule left mouse button, drag
rotate molecule (Z-axis only) Shift + right mouse button ←→
Zoom in/out Shift + left mouse button ↑↓
Move molecule Crtl + right mouse button ↑↓
Java menu right mouse button
|
| Jmol: an open-source Java viewer for chemical structures in 3D. http://www.jmol.org/
|
|

