
Elemenets of Protein Structure
Coagulation Factor VIIA Amino Acid Sequence
The purpose of this page is to demonstrate the correlation of protein sequence to protein folded structure as well as the principles that were discussed in this module using one of the essential proteins from the blood coagulation cascade as an example (factor VII).
First some background. This is a water soluble protein that is excreted in to the blood as a single folded polypeptide in an inactive state (performs no readily observable function). Upon some injury that breaks the skin, the coagulation system is set into motion. The protein, thromboplstin, is first activated by contact with platlets that have been broken open. This activated thromboplsatin performs a highly specific function, to cleave the peptide bond between ARG152 and ILE153 of factor VII converting it to the active form factor VIIa. The active form, then, has two subunits: a smaller one called the "light chain" and a larger one called the "hravy chain"
The function of factor VIIa in turn is to perform highly specific reaction, to cleave one specific peptide bond in the next factor of the blood clotting cascade (factor X converting it to factor Xa).
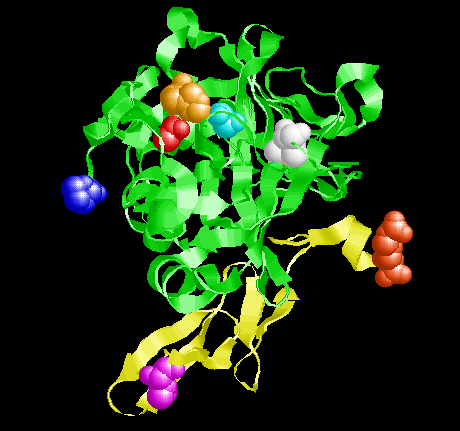
Factor VIIa (active form) with the mainchain atoms represented by the yellow (light chain) and green (heavy chain) cartoons. 6 amino acids sidechains are shown as spheres that are colored according the scheme shown in the sequence at the bottom of the page. The red amino acid attached to the yellow ribbon (Arg152) shows where factor VII is cleaved to convert to factor VIIa. in the heavy chain are shown His 57 (orange), Asp102 (red) and Ser195 (cyan).
The sequence below is the the one found in the protein expressed in humans. Every protein molecule in the human body that has this sequnce of amino acids always yields the same specific three dimensional protein structure that has the function associated Factor VIIa.
The amino acids colored in the sequence below are shown colored in the picture to the left in the same colors.
The ribbon representing the backbone of the light chain is yellow. The ribbon representing the backbone of the heavy chain is green. The side chain and backbone atoms of the first and last amino acid of each chain is colored and drawn as speheres. The three colored amino acids in the middle of the picture are three of the "critical" active site amino acids.
Structures: Factor VII - Factor VIIa Conversion
Factor VII is an inactive form for which a single peptde bond cleave leds to an active enzyme. In the literature, these are referred to as PROenzymes. This protein is initially produced as a single polypeptide inside cells it must then be excreted out to the blood. The signal that for excretion is a leader peptide (a short portion of peptide at the N-term end of a protein) that gets removed as the proteins goes through the membrane and leaves the cell. While the leader peptide is still attached, the protein is referred to as a PREprotein. Factor VII has both of these characteristics, therefore the protein initially synthesized in the cell is referred to as a PRE-PROprotein. Once in the blood, the place where factor VII is cleaved to begin the conversion to an active is between an Arg and an Ile. The paper referenced here described what was know about this process in 1986 when it was published. I do not expect you to understand all the information regarding methods, but the general idea is still apparent.
Later, in 1998, the 3D structure of the active factor VIIa was solved and then in 2001 portions of the inactive factor VII were shown. This paper shows some of the changes that occur when factor VII (inactive) is converted to factor VIIa (active) and describes some of the important events. Again the methods are beyond the scope of this course, but you can get a sense of the important issues.
Why does the protein structure rearrange upon cleavage of one peptide bond?
As shown in the picture above where the strucutures of the inactive and active proteins are superimposed, there is some slight re"settling" of the structure after the peptide bond between the Arg and Ile is broken. There is not a wholesale change of strcuture, but rather some more sublte changes. Most of the same interactions are observed between the two proteins. Howevr, as indicated earlier - nature is lazy. The protein "wants" to get to the lowest energy state possible. Once the peptide bond is broken a slightly lower energy structure is available for the protein to acquire. This is kind of like a bundle of small branches tied tightly with a string. Once you place the branches in a pile and tie them tightly with string, they have a certain arrangement in that pile. If you later cut the string, the tension is released and the branches push away from each other a bit. but the overall arrangement remains the same.... the system just relaxed to a lower energy state.
Factor VIIa Sequence*
Light Chain
ILE CYS VAL ASN GLU ASN GLY GLY CYS GLU GLN TYR CYS SER ASP HIS THR GLY THR LYS ARG SER CYS ARG CYS HIS GLU GLY TYR SER LEU LEU ALA ASP GLY VAL SER CYS THR PRO THR VAL GLU TYR PRO CYS GLY LYS ILE PRO ILE LEU GLU LYS ARG
Heavy Chain
ILE VAL GLY GLY LYS VAL CYS PRO LYS GLY GLU CYS PRO TRP GLN VAL LEU LEU LEU VAL ASN GLY ALA GLN LEU CYS GLY GLY THR LEU ILE ASN THR ILE TRP VAL VAL SER ALA ALA HIS CYS PHE ASP LYS ILE LYS ASN TRP ARG ASN LEU ILE ALA VAL LEU GLY GLU HIS ASP LEU SER GLU HIS ASP GLY ASP GLU GLN SER ARG ARG VAL ALA GLN VAL ILE ILE PRO SER THR TYR VAL PRO GLY THR THR ASN HIS ASP ILE ALA LEU LEU ARG LEU HIS GLN PRO VAL VAL LEU THR ASP HIS VAL VAL PRO LEU CYS LEU PRO GLU ARG THR PHE SER GLU ARG THR LEU ALA PHE VAL ARG PHE SER LEU VAL SER GLY TRP GLY GLN LEU LEU ASP ARG GLY ALA THR ALA LEU GLU LEU MET VAL LEU ASN VAL PRO ARG LEU MET THR GLN ASP CYS LEU GLN GLN SER ARG LYS VAL GLY ASP SER PRO ASN ILE THR GLU TYR MET PHE CYS ALA GLY TYR SER ASP GLY SER LYS ASP SER CYS LYS GLY ASP SER GLY GLY PRO HIS ALA THR HIS TYR ARG GLY THR TRP TYR LEU THR GLY ILE VAL SER TRP GLY GLN GLY CYS ALA THR VAL GLY HIS PHE GLY VAL TYR THR ARG VAL SER GLN TYR ILE GLU TRP LEU GLN LYS LEU MET ARG SER GLU PRO ARG PRO GLY VAL LEU LEU ARG ALA PRO PHE PRO
* The sequence numbers do not quite line up in this protein. For instance, it does not appear that ARG152 is the the 152nd amino acid in the sequence. Neither is His 57 the 57th. It was decided that the numbering scheme should be consistent for all proteases of this class. If you look at Trypsin, Chymotrypsin and other "serine" proteases like this one, they all have the same catalytic system - ASP-HIS-SER and they are always numbers 102, 57, 195 regardless of how many amino acids the protein has.
| | |
Shows the structure of Factor VIIa. In the two columns below, first Select a portion of the protein to highlight. Then select what you want to see and how you want to see it. Everytime you select a portion, the display items must be re-selected to activate. The cartoon of the selected portion of the protein turns red as you select it.
|
|
|
|

Click an atom to diplay it's identity here
|
|
Messages about the currently highlighted features
|
rotate molecule left mouse button, drag
rotate molecule (Z-axis only) Shift + right mouse button ←→
Zoom in/out Shift + left mouse button ↑↓
Move molecule Crtl + right mouse button ↑↓
Java menu right mouse button
| Jmol: an open-source Java viewer for chemical structures in 3D. http://www.jmol.org/
|
|

