 Enolase Information
Enolase Information Enolase Information
Enolase Information
|
Enzyme Name |
Enolase |
|
|
|
||
|
Reaction Catalyzed |
Elimination of water from 2-Phosphoglycerate
|
|
|
Reaction Type |
Elimination Reaction |
|
|
Rationale |
Elmination Reaction. The phosphate has already been placed on C2
of glycerate from the previous enzyme. The Enol functional group. The significance of the Enol group
In phosphoENOLpyruvate the enol does not terminate with -H but
with -PO3 like compound "B" Thermodynamically speaking hydrolysis of phosphate from
phosphoenolpyruvate has a very high favorable Standard Free Energy
because two things happen... we get the hydrolysis AND then the enol
converts mostly to the much more stable ketone. |
|
|
Pathway Involvement |
Glycolysis AND gluconeogenesis |
|
|
Cofactors/Cosubstrates |
a metal ion usually Mg2+ is
required as a cofactor |
|
|
|
||
|
DGo' |
+1.8 kJ/M |
Starting from standard state and allowing the reaction to come to equilibrium the 2-phosphoglycerate concentration would end up ~2 times higher than that of phosphoenolpyruvate (PEP) The Standard Free Energy favors 2-phosphglycerate. |
|
Keq |
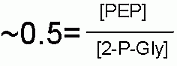 |
|
|
Comments |
|
|
|
"In cell" Substrate Concentrations* |
||
|
S1 = |
2-PhosphoGlycerate | 0.030 mM |
|
S2 = |
|
|
|
P1 = |
Phosphoenolpyruvate(PEP) | 0.023 mM |
|
P2 = |
Glyceraldehyde-3-Phosphate (G-3-P) |
0.019 mM |
|
DG for these conditions |
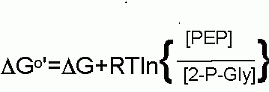 |
|
|
+1.1 kJ/M |
||
|
|
||
|
Mechanism for Chemistry |
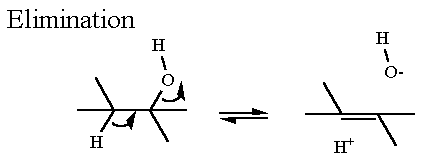 |
|
|
Mechanism for Enzyme |
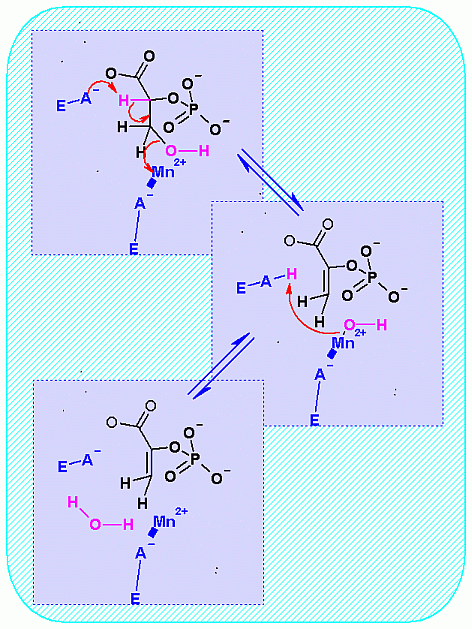
|
|
|
Picture of Enzyme with substrate |
|
|
|
|
|
|
|
Enolase CHIME representation |
|
*= These are concentrations obtained for one set of conditions. These will change as physiology and activity change.